Exploring Viral Roles in Microbial Dynamics and Carbon Cycling in the Sargasso Sea

A study published in Nature Communications utilized advanced sequencing technologies, combining short and long-read sequencing, to examine the population of viruses that infect and replicate only in bacterial cells, known as phage populations. Researchers examined them at two depths: 80 meters, the observed maximum viral concentration, and 200 meters, the mesopelagic zone beyond which light intensity rapidly dissipates as depth increases.
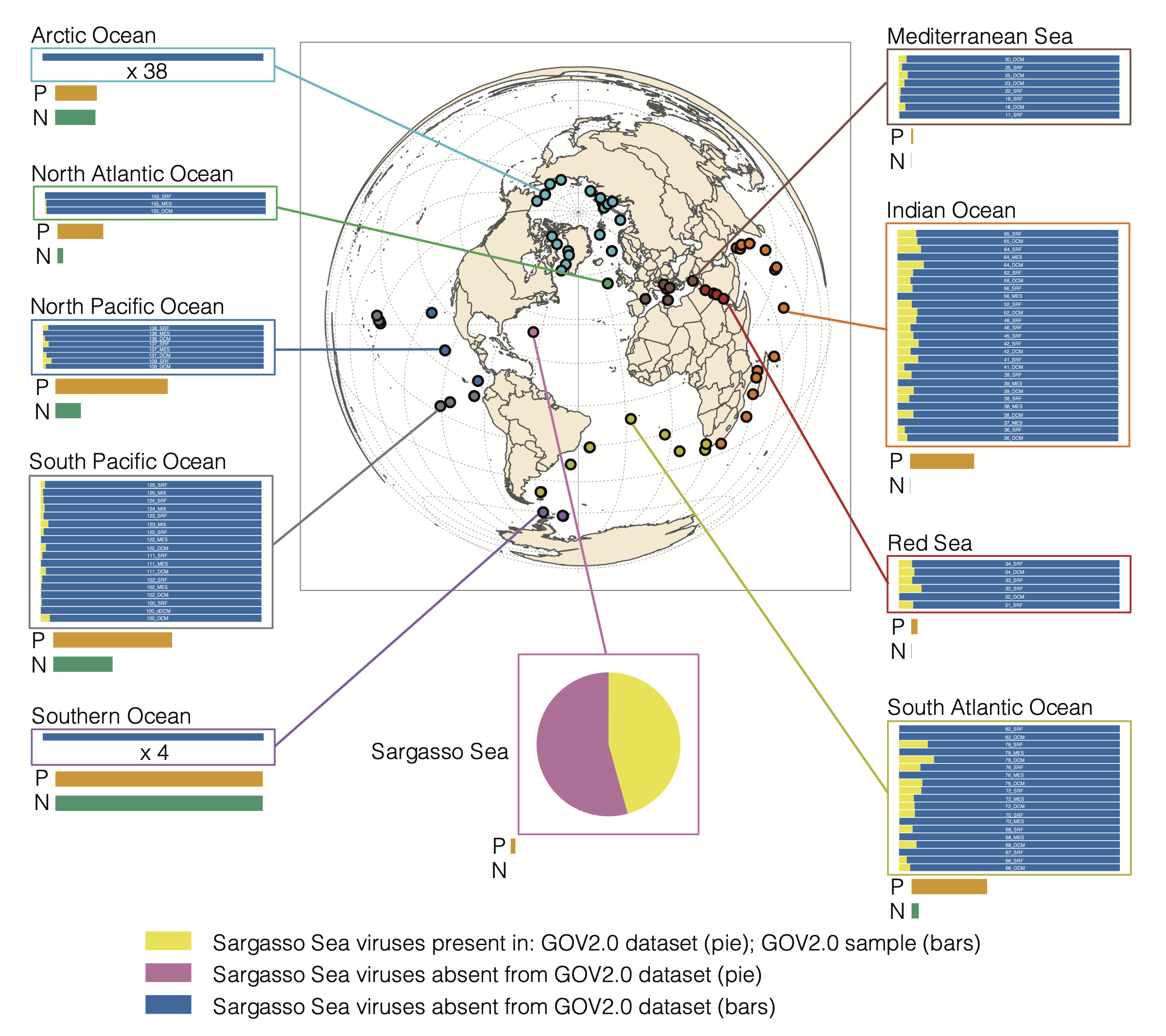
Researchers conducted a comprehensive survey of viral communities in the Sargasso Sea, a region known for its low nutrient levels, called an oligotrophic oceanic region. The Sargasso Sea is located in the Atlantic Ocean, bounded by four currents forming an ocean gyre, but unlike other seas, it has no land boundaries. This unique region is characterized by its distinctive brown Sargassum seaweed and typically calm, blue waters.
The study's key findings included the identification of 2,301 distinct phage populations from 186 genera, with over half of these phages not previously represented in global ocean viral databases; this highlights the unique viral diversity of the Sargasso Sea.
The research also revealed a significant disconnect between viral communities in the viral fraction and those viruses associated with cells, suggesting that viral turnover in this region occurs over periods longer than the three-day sampling period.
Notably, phages that infect the dominant bacterial taxa, Prochlorococcus and SAR11, were poorly represented in the viral samples, challenging previous assumptions about the role of these viruses in carbon cycling and microbial community dynamics in the Sargasso Sea.
The inclusion of long-read sequencing was crucial for capturing the full diversity of these viral populations, providing deeper insights into the ecological roles and evolutionary dynamics of marine viruses.
Lead authors of the study are Joanna Warwick-Dugdale in the School of Biosciences at the University of Exeter and Plymouth Marine Laboratory in the U.K., and Ph.D. student Funing Tian in the Center of Microbiome Science and the Department of Microbiology at The Ohio State University. Other authors include Byrd Center Principal Investigator and microbiology Professor Matthew Sullivan, director of the Center of Microbiome Science.
This study enhances our understanding of the intricate relationships between viruses and their microbial hosts in one of the most nutrient-depleted regions of the world's oceans.
Learn more about the study by visiting "Long-read powered viral metagenomics in the oligotrophic Sargasso Sea."
